Oh my goodness! Unless you are a Tree of Life developer,
you really shouldn't be here. This page is part of our beta test site, where we
develop new features for the ToL, often messing up a thing or two in the
process. Please visit the official version of this page, which is available
here.
Paleognath Relationships
John Harshman
Because the relationships presented on the Palaeognathae branch page are new and guaranteed to be controversial, it seems appropriate to show the main alternative hypotheses. For arguments why the tree on the branch page should be preferred to these alternatives, see Harshman et al. (2008).
Recent analyses of mitochondrial data have produced this tree:

Click on an image to view larger version & data in a new window
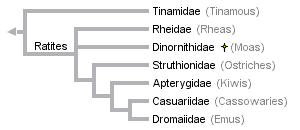
Note that ratites (all groups except the tinamous) are monophyletic. Two published analyses of nearly the same data disagree on the position of the extinct moas, and this disagreement results from the method of analysis alone. Haddrath and Baker (2001) put moas as the sister group of all other ratites, while Cooper et al (2001) put rheas as the sister group of all other ratites. The trees were otherwise identical. As in the nuclear DNA tree, there is a clade of Australasian ratites (kiwis, cassowaries, and emus).
Still more recent analyses of the same mitochondrial data, using more complex models, have produced a tree in which ratites are polyphyletic, with the relationships shown in the nuclear DNA tree (Phillips et al., 2010); in the new mitochondrial tree, tinamous are strongly supported as the sister group of moas, and those together are weakly supported as the sister group of the Australasian ratites.
A recent analysis of morphological data has produced this tree:

Click on an image to view larger version & data in a new window
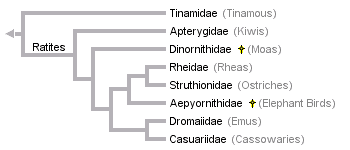
Again, note that ratites are monophyletic on this tree. The only areas of agreement with the mitochondrial tree are ratite monophyly (but see Phillips et al., 2010) and the sister relationships between emus and cassowaries (on which all trees agree). In this analysis, of Livezey and Zusi (2007), kiwis are the sister group of all other ratites, and there is no Australasian ratite clade. This is the only analysis to include elephant birds.
References
Cooper, A., C. Lalueza-Fox, S. Anderson, A. Rambaut, J. Austin, and R. Ward. 2001. Complete mitochondrial genome sequences of two extinct moas clarify ratite evolution. Nature 409:704-707.
Haddrath, O., and A. J. Baker. 2001. Complete mitochondrial DNA genome sequences of extinct birds: Ratite phylogenetics and the vicariance biogeography hypothesis. Proc. R. Soc. Lond. B 268:939-945.
Harshman, J., E. L. Braun, M. J. Braun, C. J. Huddleston, R. C. K. Bowie, J. L. Chojnowski, S. J. Hackett, K.-L. Han, R. T. Kimball, B. D. Marks, K. J. Miglia, W. S. Moore, S. Reddy, F. H. Sheldon, D. W. Steadman, S. J. Steppan, C. C. Witt, and T. Yuri. 2008. Phylogenomic evidence for multiple losses of flight in ratite birds. Proc. Natl. Acad. Sci. USA 105:13462-12467.
Livezey, B. C., and R. L. Zusi. 2007. Higher-order phylogeny of modern birds (Theropoda, Aves: Neornithes) based on comparative anatomy. II. Analysis and discussion. Zool. J. Linn. Soc. 149:1-95.
Phillips, M. J., G. C. Gibbs, E. A. Crimp, and D. Penny. 2010. Tinamous and moas flock together: Mitochondrial genome sequence analysis reveals independent losses of flight among ratites. Syst. Biol. 59:90-107.
About This Page

Correspondence regarding this page should be directed to John Harshman at
Page copyright © 2008
 Page: Tree of Life
Paleognath Relationships
Authored by
John Harshman.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Paleognath Relationships
Authored by
John Harshman.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.





 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site