Trypanosomatida
Julius Lukes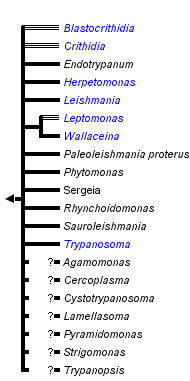

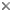
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxRelationships after Simpson et al. 2006.
Introduction
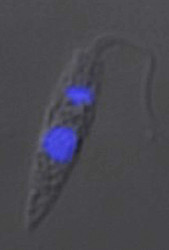
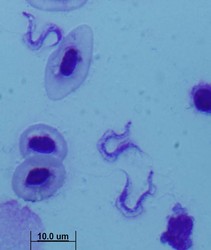
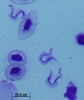
All extracellular trypanosomatid flagellates have a rather characteristic morphology of prolonged to very slender cells, twisting and actively moving by means of a single flagellum (corresponding to the anterior flagellum of bodonids), which usually exceeds the cell in size (ranging from a few microns to 100 microns). As in bodonids, the flagellum is supported by a prominent paraflagellar rod, and is usually attached to the cell by a well-visible undulating membrane. A single, usually reticulated mitochondrion contains relatively few small cristae and a kinetoplast (k) DNA invariably compacted into a single disk. The disk is composed of mutually interlocked maxicircles and minicircles and is located in the so-called paraflagellar position between the basal body of the flagellum and the nucleus. The mutual position of the kinetoplast and the nucleus, as well as the position of the flagellar pocket are variable and were used as an important taxonomic character (see below). On the ultrastructural level, salient features of trypanosomatids are inconspicuous vesicles in their cytoplasm called glycosomes. Typical Golgi apparatus and endoplasmic reticulum are present as well as a single, usually oval and centrally located nucleus containing a single nucleolus. A complete set of regularly spaced subpellicular microtubules, which stretch from the anterior to the posterior end can been seen only by electron microscopy (Vickerman 1974).
A characteristic feature of trypanosomatids is polymorphy, the ability to change their morphology during their life cycle. The main morphs of trypanosomatids are:
- amastigote – round cell with a very short flagellum not extended out of the flagellar pocket
- trypomastigote – elongated cell with the basal body and kinetoplast located at the posterior end of the cell, behind the nucleus. The flagellum emerges from a very short flagellar pocket and runs along the cell, with which it is attached via a prominent undulating membrane. The tip of the flagellum may or may not be free.
- promastigote – slender cell with the flagellum arising from the anterior tip of the cell. The kinetoplast is close to the anterior end of the cell, distant from the nucleus
- epimastigote – slender to thin cell, in which the basal body of the flagellum and the kinetoplast are located adjacent to the nucleus. The flagellum emerges from a narrow flagellar pocket in (or close to) the middle of the cell and usually forms a short undulating membrane before becoming a free flagellum
- opisthomastigote – a rare stage, characterized by a flagellum that runs from the posterior end in a very long flagellar pocket throughout the cell, emerging only from the anterior end
- choanomastigote – a rare stage, for which a rim of the flagellar pocket stretched out like a collar is typical
Presence of individual stages and the combinations thereof during the life cycle have been used for decades as genus-specific features. However, at present all genera defined in this way appear as polyphyletic in molecular studies. Moreover, there appears to be almost a continuum between these morphs, and on top of that, some of them may not appear under cultivation conditions, although they are present in the host. Therefore, it is becoming increasingly obvious that the morphology of trypanosomatids cannot be, with rather few exceptions, used as the basis for a taxonomic system (Yurchenko et al., 2006; 2008).
Characteristics
Trypanosomatid flagellates of the genera Trypanosoma and Leishmania undoubtedly belong to the most studied protists. This is not only because they are causative agents of serious human diseases, but also because they have an exceptionally high number of unique features. In some cases, the departure of the kinetoplastid cell from a regular eukaryotic cell is stunning. It is also worth mentioning that some trypanosomatids, such as Trypanosoma brucei, Leishmania tarentolae and Crithidia fasciculata represent model species that are amenable to a number of molecular biology methods, can be cultivated in cheap media at high density, and do not pose any risk for the investigator. While in all three species genes can be conveniently knocked-out by homologous recombination, downregulation of transcripts by RNA interference is so far operational only in T. brucei. The complete genome has been sequenced for T. brucei, T. cruzi and L. major (Berriman et al., 2005), but efforts to sequence the genomes of several other members of the genera Leishmania and Trypanosoma are under way. A eukaryotic expression system is being developed for L. tarentolae that seems to be particularly useful for the overexpression of human proteins (Breitling et al., 2002).
The list of unique features of the trypanosomatid cell is long, so only the best known oddities will be briefly mentioned here. Kinetoplast (k) DNA in the single mitochondrion of trypanosomatids has a unique structure, being composed of thousands of mutually interlocked minicircles and dozens of maxicircles (Lukes et al., 2005). Perhaps hundred of proteins are required for maintenance and replication of this extremely complicated network. Maxicircles encode homologues of typical mitochondrial genes of other eukaryotes, however, some of them are present in an encrypted form, which means that their transcripts have to undergo RNA editing by the insertions and deletions of uridines in order to become translatable on mitochondrial ribosomes. Information from the exact editing is specified by hundreds of small RNA molecules termed guide RNAs that are encoded by minicircles. Moreover, dozens of proteins, constituting several unique protein complexes, are necessary for the process. It is worth mentioning here that due to its massive presence in kinetoplastid flagellates, facilitating its seminal discovery (Benne et al., 1986), RNA editing was since found in virtually all groups of organisms and belongs to the key mechanisms of RNA metabolism.
Another unusual feature of the trypanosomatid cell is the presence of glycosomes in its cytosol. Glycosomes are organelles most likely derived from peroxisomes. Part of the glycolytic pathway (the first seven enzymes) is sequestered in these membrane-bound organelles, which do not possess any DNA. The bloodstream stages of trypanosomes use glucose as the sole source of carbon and energy, as they have virtually unlimited access to glucose in the host’s blood. It is metabolized by a form of aerobic respiration at an extremely high rate, which is necessary to support metabolism of the parasite, as the end-product is pyruvate and only little ATP is produced in the process. This is mainly due to the lack of the classical tricarboxylic (Krebs) cycle and standard respiratory chain.
On the level of RNA metabolism, two features should be mentioned that are both either absent or rare in eukaryotes, but are present in a massive form in trypanosomatids. In their nucleus, protein-encoding genes are transcribed polycistronically (Campbell et al., 2003). The huge polycistronic precursors are excised into individual mRNAs, and a short stretch of 39 nucleotides, termed spliced leader (SL RNA) is attached to the 5’ end of each and every mRNA via trans-splicing. In addition to typical methylation of the cap, several additional bases of the SL RNA are methylated, resulting in a hypermethylated cap. The available data indicates that there is little if any transcription regulation and that most regulation occurs at the post-transcriptional level, representing another oddity of these ancient parasites (Gunzl 2003).
Last but definitely not least, a unique feature of the African trypanosomes is antigenic variation. The surface coat of an individual cell consists of several million molecules of a single molecular species of variant surface glycoprotein (VSG). VSG exists in the form of a dimer, constituting a macromolecular diffusion barrier. Antigenic variation involves the sequential expression of coats composed of different VSGs (Cross 2003). The changing surface coat protects the parasite from the immune response of the host. The purification of abundant VSG allowed the identification of the glycosylphosphatidylinositol (GPI) anchor in trypanosomes, which was later found to be widespread in eukaryotic cells. There are several hundred VSG genes in the T. brucei genome, and new versions of the gene are being generated by recombination. The expression of VSG can be changed by turning off one expression site and turning on another one. This unparalleled capacity of African trypanosomes to change their surface coat practically eliminates the chances for a successful vaccine (Cross 2003).
Habitat and Pathogenicity
Trypanosomatida include facultatively parasitic species that are either pathogenic dixenous members, or monoxenous species restricted to invertebrate hosts. The two most important genera are Trypanosoma and Leishmania, the members of which are responsible for numerous diseases of humans and animals. The most important disease is African sleeping sickness caused by T. brucei transmitted to humans by the tsetse fly (Glossina spp.). An even more widespread disease confined to Central and South America is Chagas disease caused by T. cruzi and transmitted by the triatomine bugs (e.g. Rhodnius spp.). However, Trypanosoma species have been found in virtually every vertebrate host, being particularly widespread in fish and birds.
Leishmaniasis is present in most tropical countries, with hundreds of thousands of cases reported annually from Asia and East Africa. The disease is transmitted by phlebotomine sandflies that transmit more than a dozen of species pathogenic for humans. Main forms are cutaneous leishmaniasis, which may be mutilating and disfiguring and visceral leishmaniasis, which is systemic and mostly lethal if left untreated (Lukes et al., 2007). Members of the digenetic genus Phytomonas are transmitted by insects and are associated with severe pathological syndromes of plants (see below).
The trypanosomatids include highly pathogenic species with extracellular (T. brucei) or intracellular localization (T. cruzi), some being confined within the host cell to the parasitophorous vacuole (Leishmania spp.). Some species are remarkably non-pathogenic, such as T. rangeli.
Important Diseases Caused by Trypanosomatids
Trypanosoma spp.
| Species/subspecies | Disease | Hosts | Distribution |
|---|---|---|---|
| T. brucei brucei | nagana (n’gana) | cattle, horses, wild game | Africa |
| T. brucei rhodesiensis | sleeping sickness (severe) | humans, pigs, wild game | Africa |
| T. brucei gambiensis | sleeping sickness (mild) | humans, wild game | Africa |
| T. congolense | trypanosomiasis | cattle, wild game | Africa |
| T. cruzi | Chagas disease | humans, wild animals | South and Central America |
| T. brucei evansi | surra | horses, camels and water buffaloes | South and Central America, Asia, Europe, Africa |
| T. brucei equiperdum | dourine | horses, donkeys, camels | South and Central America, Asia, Europe, Africa |
Leishmania spp.
| Species | Disease | Hosts | Distribution |
|---|---|---|---|
| L. braziliensis | espundia, mucocutaneous leishmaniasis | humans, rodents | South America |
| L. mexicana | kala azar, visceral leishmaniasis | humans, rodents | Central America |
| L. donovani | sleeping sickness (mild) | humans, dogs | Africa, Asia, Europe |
| L. major | cutaneous leishmaniasis (wet form) | humans, rodents | Africa, Asia |
| L. tropica | cutaneous leishmaniasis (dry form) | humans, hyrax | Africa, Asia, Europe |
| L. infantum | visceral leishmaniasis (mostly) | humans, dogs | Africa, Europe, South America |
| Phytomonas spp. | hartrot disease | coconut, oil palms | Central and South America |
L. chagasi = synonymized recently with L. infantum
References
Benne, R., J. Van den Burg, J.P.J. Brakenhoff, P. Sloof , J.H. Van Boom and M.C. Tromp. 1986. Major transcript of the frameshifted coxII gene from trypanosome mitochondria contains 4 nucleotides that are not encoded in the DNA. Cell 46: 819-826.
Berriman M., E. Ghedin and C. Hertz-Fowler. 2005. The genome of the African trypanosome Trypanosoma brucei. Science 309: 416-422.
Breitling R., S. Klinger and N. Callewaert. 2002. Non-pathogenic trypanosomatid protozoa as a platform for protein research and production. Protein Expr. Purif. 25: 209-218.
Brun R. and O. Balmer. 2006. New development in human African trypanosomiasis. Curr. Opinion Infect. Dis. 19: 415-420.
Campbell D.A., S. Thomas and N.R. Sturm. 2003. Transcription in kinetoplastid protozoa: why be normal. Microbes and Infection 5: 1231-1240.
Cross G.A.M. 2003. Antigenic variation in African trypanosomes and malaria. In: Molecular Medical Parasitology (edited by Marr J.J., Nilsen T.W. & Komuniecki R.W.).
De Souza W. 2007. Chagas disease: facts and reality. Microbes Infect. 9: 544-545.
De Souza W. and M.C.M. Motta. 1999. Endosymbiosis in protozoa of the Trypanosomatidae family. FEMS Microbiol. Lett. 173: 1-8.
Dujardin J.C. 2006. Risk factors in the spread of leishmanises: towards integrated monitoring? Trends Parasitol. 22: 4-6.
Gunzl A. 2003. Transcription. In: Molecular Medical Parasitology (edited by Marr J.J., Nilsen T.W. & Komuniecki R.W.).
Lai D.H., H. Hashimi, Z. Lun, F.J. Ayala and J. Lukeš. 2008. Adaptation of Trypanosoma brucei to gradual loss of kinetoplast DNA: T. equiperdum and T. evansi are petite mutants of T. brucei. Proc. Natl. Acad. Sci. USA 105: 1999-2004.
Lukeš J., H. Hashimi and A. Zikova. 2005. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet. 48: 277-299.
Lukeš J., I.L. Mauricio, G. Schönian, J.C. Dujardin, K. Soteriadou, J.P. Dedet, K. Kuhls, W. Quispe Tintaya, M. Jirků, E. Chocholová, C. Haralambous, F. Pratlong, M. Oborník, A. Horák, F.J. Ayala and M.A. Miles. 2007. Evolutionary and geographical history of the Leishmania donovani complex with a revision of current taxonomy. Proc. Natl. Acad. Sci. USA 104: 9375-9380.
Maslov D.A. and L. Simpson. 1994. RNA editing and mitochondrial genomic organization in the cryptobiid kinetoplastid protozoan Trypanoplasma borreli. Mol. Cell. Biol. 14: 8174-8182.
Schonian G., A. Nasereddin, N. Dinse, C. Schweynoch, H.D.F.H Schallig, W. Presber and C.L. Jaffe. 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis. 47: 349-358.
Simpson A.G.B., J.R. Stevens and J. Lukeš. 2006. The evolution and diversity of kinetoplastid flagellates. Trends Parasitol. 22: 168-174.
Vickerman K. 1974. The ultrastructure of pathogenic flagellates. Trypanosomiasis and Leishmaniasis. Elsevier.
Westenberger S.J., C. Barnabe, D.A. Camnpbell and N.R. Sturm. 2005. Two hybridization events define the population structure of Trypanosoma cruzi. Genetics 171: 527-543.
Yurchenko V., J. Lukeš, X. Xu and D.A. Maslov. 2006. An integrated morphological and molecular approach to a new species description in the Trypanosomatidae: the case of Leptomonas podlipaevi n.sp., a parasite of Boisea rubrolineata (Hemiptera: Rhopalidae). J. Euk. Microbiol. 53: 103-111.
Yurchenko V.A., J. Lukeš, M. Tesařová, M. Jirků and D.A. Maslov. 2008. Morphological discordance of the new trypanosomatid species phylogenetically associated with the genus Crithidia. Protist 159: 99-114.
Title Illustrations

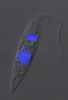
| Scientific Name | Leishmania major |
|---|---|
| Comments | from Promastigote (culture form) |
| Specimen Condition | Live Specimen |
| Copyright |
© 2008 Jan Votýpka

|
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.

University of South Bohemia in Ceske Budejovice, Czech Republic
Correspondence regarding this page should be directed to Julius Lukes at
Page copyright © 2009
All Rights Reserved.
- First online 02 January 2009
- Content changed 02 January 2009
Citing this page:
Lukes, Julius. 2009. Trypanosomatida . Version 02 January 2009 (under construction). http://tolweb.org/Trypanosomatida/98015/2009.01.02 in The Tree of Life Web Project, http://tolweb.org/





 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site