Plasmodium
Malaria Parasites
Martine Zilversmit and Susan Perkins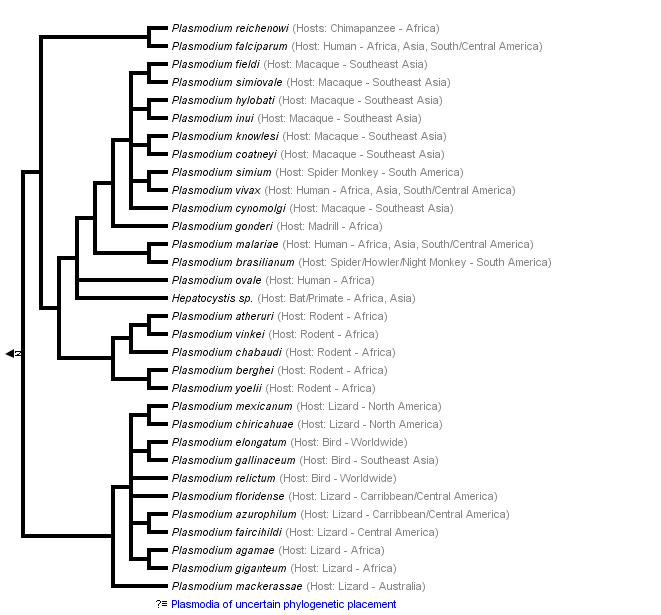

This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
Organisms that belong to the genus Plasmodium are obligate eukaryotic parasites, best known as the etiological agent of human malaria. There are four parasites that infect humans and cause malaria: P. falciparum, P. vivax, P. malariae, and P. ovale. Although Plasmodium parasites infect a variety of vertebrate hosts (including primates, rodents, ungulates, birds, and lizards), they rarely cause severe disease in any vertebrate hosts other than humans. The most virulent of human parasites, P. falciparum and P. vivax (White, 2003), cause 300-500 million cases of debilitating or fatal disease worldwide.
Our understanding of the evolution and systematics of malaria parasites has changed significantly over the past 20 years with recent work (Perkins and Schall, 2002; Martinsen et al., 2008) indicating that the genus Plasmodium may not to be monophyletic, and includes parasites of other genera including Hepatocystis.
Characteristics
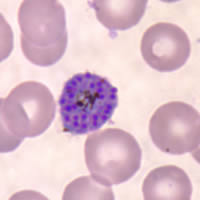
Organisms of the genus Plasmodium are defined as distinct from other Apicomplexa, and other organisms sometimes considered malaria parasites (Peréz-Tris et al. 2005), as parasites that both undergo merogony (multiple divisions of the nucleus followed by segmentation of the cytoplasm producing daughter cells called merozoites) in erythrocytes, and that produce hemozoin pigment, the crystalline by-product of hemoglobin digestion. Other members of the order Haemosporida vary in the combination of these characters. The family Haemoproteidae produces pigment, but merogony occurs in tissues other than erythrocytes. The families Garniidae and Leucocytozoidae do not produce pigment at any stages, but in the case of the former do show merogony in blood cells.

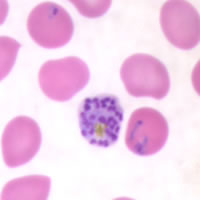
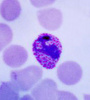
P. falciparum schizont stage parasite (center) showing merogony, recognizable by approximately 24 new merozoites identified by the Giemsa-stained nuclei (purple), and showing hemozoin pigment (yellow)
All malaria parasites have a sexual life stage that occurs in a blood-feeding insect, which is the definitive host for these organisms (also known as the “vector,” in epidemiology).
Traditionally, Plasmodium species were described based on morphological and morphometric characteristics, primarily of the blood stage of the lifecycle. Other characters used in the past for classification have been virulence-level and life-cycle period in the erythrocytic stage (24, 48, or 72 hours). Morphological characterization has often proven to be insufficient to distinguish species, and has also been shown to conflict with molecular sequence data. Scientists are beginning to use advanced methods and molecular characters for parasite taxonomy and phylogenetics. (See Discussion of Phylogenetic Relationships.)
Hosts and Geographic Distribution of Some Parasites
| Parasite | Host | Geographic Distribution |
|---|---|---|
| Plasmodium reichenowi | Chimapanzee | Africa |
| Plasmodium falciparum | Human | Africa, Asia, South/Central America |
| Plasmodium fieldi | Macaque | Southeast Asia |
| Plasmodium simiovale | Macaque | Southeast Asia |
| Plasmodium hylobati | Macaque | Southeast Asia |
| Plasmodium inui | Macaque | Southeast Asia |
| Plasmodium knowlesi | Macaque | Southeast Asia |
| Plasmodium coatneyi | Macaque | Southeast Asia |
| Plasmodium simium | Spider Monkey | South America |
| Plasmodium vivax | Human | Africa, Asia, South/Central America |
| Plasmodium cynomolgi | Macaque | Southeast Asia |
| Plasmodium gonderi | Madrill | Africa |
| Plasmodium malariae | Human | Africa, Asia, South/Central America |
| Plasmodium brasilianum | Spider/Howler/Night Monkey | South America |
| Plasmodium ovale | Human | Africa |
| Hepatocystis sp. | Bat/Primate | Africa, Asia |
| Plasmodium atheruri | Rodent | Africa |
| Plasmodium vinkei | Rodent | Africa |
| Plasmodium chabaudi | Rodent | Africa |
| Plasmodium berghei | Rodent | Africa |
| Plasmodium yoelii | Rodent | Africa |
| Plasmodium mexicanum | Lizard | North America |
| Plasmodium chiricahuae | Lizard | North America |
| Plasmodium elongatum | Bird | Worldwide |
| Plasmodium gallinaceum | Bird | Southeast Asia |
| Plasmodium relictum | Bird | Worldwide |
| Plasmodium floridense | Lizard | Carribbean/Central America |
| Plasmodium azurophilum | Lizard | Carribbean/Central America |
| Plasmodium faichildi | Lizard | Central America |
| Plasmodium agamae | Lizard | Africa |
| Plasmodium gigantum | Lizard | Africa |
| Plasmodium mackerassae | Lizard | Australia |
Plasmodium Life-cycle
Plasmodium parasites have an elaborate life-cyle with multiple stages:
- Infective stage, when the parasite enters the vertebrate host with a vector bite. This life stage is known as sporozoite.
- Exoerythrocytic stage, in which the sporozoite undergoes multiple rounds of asexual divisions (merogony or schizongony) and matures into merozoites.
- Erythrocytic stages, during which the organisms enter red blood cells (as merozoites), transform into the feeding stages (trophozoites), and then divide asexually into multiple new merozoites (schizont stage). During the schizont stage, some parasites differentiate into the reproductive forms (gametocytes) rather than the invasive merozoites. Gametocytes are classified as microgametocytes (that will become male gametes) and macrogametocytes (that will become female gametes). The gametocytes must mature through five stages before they become infective to the mosquito.
- Reproductive stages, these begin when the vector takes a blood meal from the vertebrate host that contains mature gametocytes. In the vector the gametocytes transform into male and female gametes and merge to become a zygote (the only diploid stage in the organism's life-cycle). The zygote becomes an ookinate which invades the tissues of the vector midgut to become an oocyst. When the oocyst ruptures thousands of sporozoites emerge and travel to the vector's salivary glands, as it is through the saliva that they will enter the next vertebrate host.
Discussion of Phylogenetic Relationships
The field of Plasmodium phylogenetics has been particularly dynamic over the past decade, and continues to be so as more species are added to the tree and older hypotheses are called into question.
The study of the molecular phylogenetics of malaria parasites began in 1991 (Waters et al., 1991), and the field continues to grow. The conclusions of many earlier studies were overturned later, as they were affected by difficulties stemming from insufficient taxon sampling and problems with the genes chosen for analysis. The choice of both the ingroup taxa (the organisms of interest) and outgroup taxa (organisms outside the group of interest) for phylogenetic analysis can have a significant influence on the strength of the resulting evolutionary hypothesis. Older phylogenies often contained small numbers of ingroup species, between six and twelve, that were a mixture of very closely-related taxa and a few that were very divergent. This poor taxon sampling for this very large and diverse genus, coupled with the use of very distantly-related species as outgroups, probably lead to some spurious results and the conflicting conclusions found in the literature at the time.
In addition to problems of taxon sampling, for years the study of malaria systematics was also stymied by inherent problems in the loci chosen for analysis. Small subunit (SSU) rRNA and circumsporozoite protein (CSP) have been the workhorses of Plasmodium phylogenetics (Waters et al., 1991; Waters et al., 1993; Escalate and Ayala, 1994; Escalante et al., 1995; McCutchan, et al. 1996; Qari et al., 1996; Escalante et al., 1996; Escalante et al., 1997). Recent research indicates, however, that neither of these loci may be appropriate for evolutionary studies of Plasmodium species. SSU rRNA is a standard locus used in high-level molecular systematics, but it was later found that Plasmodium species possess separate genes of rRNA, each expressed at a different point in the life cycle, that are not evolving in a concerted manner and may still be exchanging genetic information with each other (Corredor and Enea, 1993). Several of the older phylogenies may have included a mixture of paralogs (duplicate genes) and orthologs (proper homologs), and only orthologs yield reliable gene trees. Although life-stage-specific primers have been developed for the SSU rRNA loci in Plasmodium, as gene conversion among the non-homogenized paralogs cannot be ruled out, this locus should be used with great caution. Indeed, even very recent phylogenies (Leclerc et al., 2004) using these sequences show results that cannot be reconciled with those of other loci in the genome.
Likewise, the locus CSP, which has been frequently used for evolutionary studies, may be problematic as the gene codes for a surface protein and is under strong selective pressure from the vertebrate immune system. Selectively driven, non-neutral changes in the gene may either obscure the phylogenetic signal or lead to incorrect phylogenetic inferences (Hughes and Hughes, 1995). CSP was a favored locus for study as large amounts of sequence data was available early on as the protein is of great interest to malaria science as a possible vaccine target. Improved methods for gene sequencing have allowed for better loci to be developed for evolutionary studies and CSP is now rarely used in systematics.
At this time, new loci are being developed for molecular studies, such as the mitochondrial gene for cytochrome b (Escalante et al., 1998; Perkins and Schall 2002) and the gene for the housekeeping enzyme adenylosuccinate lyase (Kedseierki et al., 2002). One caveat is that mitochondrial cytochrome b is the target of some antimalarial drugs, and mutations in the gene are known to be associated with resistance (see Vaidya, et al., 1993). Studies of these and other suitably chosen genes will hopefully lead to Plasmodium phylogenies based on the combined data from multiple loci. It should be noted, however, that the chance of identifying true neutrally evolving loci in Plasmodium species is compromised by the possibility of unrecognized selection pressure from the host immune system or (in the case of human parasites) drugs, and hence an extra measure of caution is called for.
One intuitive assumption that was challenged early on by molecular studies was the idea that human malaria parasites are closely related. Recent work now shows that, although primate parasites (with the exception of P. falciparum and P. reichenowi) grouped together, those parasites with a human host were not closely related within this group. The relatively distant relationships among human pathogens indicates that malaria most likely has an ancient association with primates (Escalante et al., 1998; Perkins and Schall, 2002; Leclerc et al., 2004, Cornejo and Escalante, 2006).
One of the most intriguing issues in malaria evolution is the mysterious origin of P. falciparum, and few issues have gone through as many significant revisions in the past 15 years. Older textbooks, and even some current ones, refer to P. falciparum as arising from a recent horizontal transfer from birds, and it is to this recent host shift that the high virulence of P. falciparum is often attributed (White, 2003). Newer research, however, refutes this hypothesis, as P. falciparum falls within a clade of mammalian parasites—distinct from and outside the group affecting birds and lizards. On the other hand, the species is also outside of the primate or rodent malarias. Rather, P. falciparum is in a divergent clade that includes only itself and its close sister species, the chimpanzee parasite, P. reichenowi.
The avian-origin hypothesis arose from some of the early molecular phylogenies that had problems due to insufficient taxon sampling and choice of genes (Waters et al., 1991; Waters et al., 1993; Escalante et al., 1995; Escalante et al., 1996; Escalante et al., 1997). First in 1996 (Qari et al., 1996) and then again in 2002 (Perkins and Schall, 2002), new evidence came to light refuting an avian-origin. Perkins and Schall (2002) published a relatively strong phylogeny based on cytochrome b with a large sample size of vertebrate parasites and an outgroup from a sister family. This cytochrome b phylogeny placed P. falciparum within the clade of mammalian parasites. In addition to the phylogenetics, this analysis included an explicit test of the avian-origin hypothesis using the Shimodaira-Hasegawa test, a phylogenetic tree-based method that calculates the likelihood of alternate trees, and rejected it.
At this time, the evidence (Qari et al., 1996; Escalate and Ayala, 1997; Perkins and Schall, 2002; Leclerc et al., 2004; Martinsen et al., 2008) indicates that the only species closely related to P. falciparum is P. reichenowi, and the two likely diverged from each other between 5 and 8 million years ago based on fossil dates of the human-chimpanzee split (Escalante and Ayala, 1994; Escalante et al., 1995).
Future phylogenetic analyses that include parasites that infect bats (several genera of malaria parasites have been described from these hosts alone) and ungulates may help to identify close relatives of P. falciparum and better understand the evolutionary history of these parasites.
References
Coatney GR, Collins WE, Warren M, Contacos PG. 1971. The Primate Malarias. US Department of Health, Education and Welfare, Bethesda.
Cornejo OE, Escalante AA. 2006. The origin and age of Plasmodium vivax. Trends Parasitol. 22(12):558-63.
Corredor V, Enea V. 1993. Plasmodial ribosomal RNA as phylogenetic probe: a cautionary note. Mol. Biol. Evol. 10:924-926.
Escalante AA, Ayala FJ. 1994. Phylogeny of the malarial genus Plasmodium, derived from rRNA gene sequences. Proc. Natl. Acad. Sci. USA 91:11373-11377.
Escalante AA, Barrio E, Ayala FJ. 1995. Evolutionary origin of human and primate malarias: evidence from the circumsporozoite protein. Mol. Biol. Evol. 12:616-626.
Escalante AA, Freeland DE, Collins, WE, Lal AA. 1998. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc. Natl. Acad. Sci. USA 95:8124-8129.
Escalante AA, Goldman IF, De Rijk P, De Wachter R, Collins WE, Qari SH, Lal. AA 1997. Phylogenetic study of the genus Plasmodium based on the secondary structure-based alignment of the small subunit ribosomal RNA. Mol. Biochem. Parasitol. 90:317-321.
Hughes MK, Hughes AL. 1995. Natural selection on Plasmodium surface proteins. Mol Biochem Parasitol. 71(1):99-113.
Kedzierski L, Escalante AA, Isea R, Black CG, Barnwell JW, Coppel RL. 2002. Phylogenetic analysis of the genus Plasmodium based on the gene encoding adenylosuccinate lyase. Infect Genet Evol. 1(4):297-301.
Leclerc MC, Hugot JP, Durand P, Renaud F. Evolutionary relationships between 15 Plasmodium species from new and old world primates (including humans): an 18S rDNA cladistic analysis. 2004. Parasitology. 129(6):677-84.
Martinsen ES, Perkins SL, Schall JJ. 2008. A three-genome phylogeny of malaria parasites (Plasmodium and closely related genera): Evolution of life-history traits and host switches. Mol Phylogenet Evol 47(1):261-273.
McCutchan TF, Kissinger JC, Touray MG, Rogers MJ, Li J, Sullivan M, Braga EM, Krettli AU, Miller LH. 1996.Comparison of circumsporozoite proteins from avian and mammalian malarias: biological and phylogenetic implications. Proc Natl Acad Sci U S A. 93:11889-94.
Perez-Tris J, Hasselquist D, Hellgren O, Krizanauskiene A, Waldenstrom J, Bensch S. 2005. What are malaria parasites? Trends Parasitol. 21(5):209-11.
Perkins SL, Sarkar IN, Carter R. 2007. The phylogeny of rodent malaria parasites: simultaneous analysis across three genomes. Infect Genet Evol. 7(1):74-83.
Perkins SL, Schall JJ. 2002. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 88(5):972-8.
Qari SH, Shi YP, Pieniazek NJ, Collins WE, Lal AA. 1996. Phylogenetic Relationship among the Malaria Parasites Based on Small Subunit rRNA Gene Sequences: Monophyletic Nature of the Human Malaria Parasite, Plasmodium falciparum. Mol. Phylo. Evol. 6(1)157-165.
Vaidya AB, Lashgari MS, Pologe LG, Morrisey J. 1993. Structural features of Plasmodium cytochrome b that may underlie susceptibility to 8-aminoquinolines and hydroxynaphthoquinones. Mol Biochem Parasitol. 58(1):33-42.
Vargas-Serrato E, Corredor V, Galinski MR. 2003. Phylogenetic analysis of CSP and MSP-9 gene sequences demonstrates the close relationship of Plasmodium coatneyi to Plasmodium knowlesi. Infect. Genet. Evol. 3(1):67-73.
Waters AP, Higgins DG, McCutchan TF. 1991. Plasmodium falciparum appears to have arisen as a result of a lateral transfer between avian and human hosts. Proc. Natl. Acad. Sci. USA 88:3140-3144
Waters AP, Higgins DG, McCutchan TF. 1993. Evolutionary relatedness of some primate models of Plasmodium. Mol Biol Evol. 10(4):914-23.
White NJ. 2003. Malaria, pp. 1205-1296. In G. C. Cook, and A. Zumla (ed.), Mansonís Tropical Diseases, 21st ed. WB Saunders, Philadelphia.
Title Illustrations

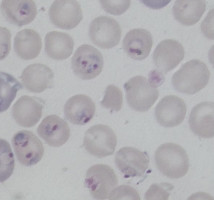
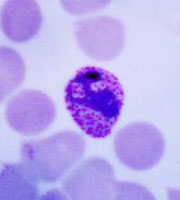
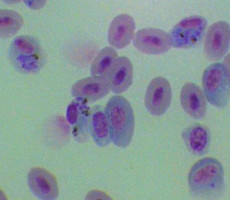
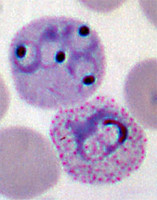
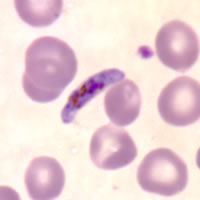
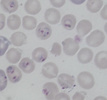
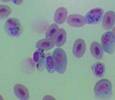
Plasmodium parasites in the erythrocytic stages: left, P. chabaudi in early trophozoite "ring" stage; P. vivax in trophozoite stage; P. cathemerium in schizont stages.
| Scientific Name | Plasmodium chabaudi |
|---|---|
| Specimen Condition | Fixed Specimen on Slide |
| Sex | Asexual |
| Life Cycle Stage | "Ring" stage/early trophozoite |
| Copyright | © 2007 Brian Culleton |
| Scientific Name | Plasmodium vivax |
|---|---|
| Specimen Condition | Fixed specimen on microscope slide |
| Sex | Asexual |
| Life Cycle Stage | Mature trophozoite |
| Source | PHIL_2725_lores.jpg |
| Source Collection | Public Health Image Library (Centers for Disease Control) |
| Scientific Name | Plasmodium cathemerium |
|---|---|
| Comments | An avian malaria parasite. See Plasmodia of uncertain phylogenetic placement |
| Specimen Condition | Fixed Specimen on Slide |
| Identified By | Thomas Buckelew |
| Sex | Asexual |
| Copyright | © 2007 Thomas Buckelew |
About This Page
Martine Zilversmit

New York University School of Medicine, New York, New York, USA
Susan Perkins

American Museum of Natural History, New York, New York, USA
Correspondence regarding this page should be directed to Martine Zilversmit at
zilversmit@nyu.edu
and Susan Perkins at
perkins@amnh.org
Page copyright © 2008 Martine Zilversmit and Susan Perkins
 Page: Tree of Life
Plasmodium. Malaria Parasites.
Authored by
Martine Zilversmit and Susan Perkins.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial-ShareAlike License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Plasmodium. Malaria Parasites.
Authored by
Martine Zilversmit and Susan Perkins.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial-ShareAlike License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
- First online 24 April 2008
- Content changed 05 May 2008
Citing this page:
Zilversmit, Martine and Susan Perkins. 2008. Plasmodium. Malaria Parasites. Version 05 May 2008. http://tolweb.org/Plasmodium/68071/2008.05.05 in The Tree of Life Web Project, http://tolweb.org/





 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site