Hacrobia
Patrick Keeling

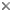
This tree diagram shows the relationships between several groups of organisms.
The root of the current tree connects the organisms featured in this tree to their containing group and the rest of the Tree of Life. The basal branching point in the tree represents the ancestor of the other groups in the tree. This ancestor diversified over time into several descendent subgroups, which are represented as internal nodes and terminal taxa to the right.

You can click on the root to travel down the Tree of Life all the way to the root of all Life, and you can click on the names of descendent subgroups to travel up the Tree of Life all the way to individual species.
For more information on ToL tree formatting, please see Interpreting the Tree or Classification. To learn more about phylogenetic trees, please visit our Phylogenetic Biology pages.
close boxIntroduction
The Hacrobia is a large group comprising a great diversity of microbial eukaryotes, including flagellates, amoebae, microalgae, and heterotrophs. Two of the subgroups of Hacrobia, cryptomonads and haptophytes are large and well studied lineages of important primary producers, while kataplepharids, centrohelid heliozoa and telonemids are all abundant heterotrophs, primarily in the marine environment. The group was only recently proposed and is based exclusively on molecular data since no single character that unites these groups on the basis of morphology has been identified.
Discussion of Phylogenetic Relationships
Historically the various subgroups of Hacrobia have been classified in many different ways, but generally they were not considered to be related to one another until relatively recently. The cryptomonads and haptophytes were both proposed to be part of the Chromalveolates, but not as sisters within that group (Cavalier-Smith 1999). Katablepharids have also been classified with cryptomonads based on similarities observed from light microscopy, but they were eventually re-classified as incertae sedis based on ultrastructure (Vørs 1992).
From molecular data, several hacrobian subgroups were proposed to be related to one another. Phylogenetic analyses based on LSU and SSU rDNA confirmed that katablepharids are closely related to cryptophytes (Okamoto and Inouye 2005; Kim, Simpson, and Graham 2006; Okamoto and Inouye 2006). This has since been supported by analyses of additional genes (Kim, Simpson, and Graham 2006; Okamoto et al. 2009). Biliphytes were also found to be closely related to cryptophytes based on phylogeny of SSU rDNA (Not et al. 2007; Cuvelier et al. 2008; Vaulot et al. 2008; Not et al. 2009), but this enigmatic lineage is known only from environmental sequences and fluorescence in situ hybridization (FISH). The phylogenetic position of telonemids based on different genes has been more variable (Klaveness et al. 2005; Shalchian-Tabrizi et al. 2006; Shalchian-Tabrizi et al. 2007), but one analysis of HSP90 and a small concatenation of genes suggested some relationship to cryptomonads (Shalchian-Tabrizi et al. 2006). The centrohelid heliozoans, which are now known not to be closely related to other members of the traditional Heliozoa (Nikolaev et al. 2004; Sakaguchi et al. 2005) were not clearly placed in molecular trees for quite some time, although a weakly supported SSU rDNA tree and some intriguing ultrastructural similarities led to the suggestion that they were related to haptophytes (Cavalier-Smith and von der Heyden 2007).
More definitive data for most of these groups have come from analysis of large collections of nuclear genes. These multigene analyses first indicated a close relationship between haptophytes and cryptomonads (Hackett et al. 2007; Patron, Inagaki, and Keeling 2007), which has since been confirmed in other large scale analsyses (Hampl et al. 2009). Following this, similar analyses including telonemids and centrohelids also found these two groups to be related to cryptomonads and haptophytes, although not with as strong support and not in a clearly definable position within the growing Hacrobia group (Burki et al. 2009).
A significant shared molecular feature was also identified in the plastid genome of haptophytes and cryptomonads. Horizontal gene transfer to plastid genomes is very rare, but both haptophyte and cryptomonad plastid genomes were found to have acquired the same copy of the bacterial gene for rpl36, which evidently replaced the existing gene (Rice and Palmer 2006). No other plastid has this features, making this a strong indication that haptophyte and cryptomonad plastids share a common ancestor. This, together with the nuclear gene data supporting the relationship between the nuclear lineages of various hacrobian subgroups, suggests the Hacrobia all derived from a common ancestor and that this ancestor was a photosynthetic alga that contained a plastid derived from secondary endosymbiosis with a red alga (Keeling 2009). Based on this, the Hacrobia was formally proposed, the name deriving from the names of the two largest subgroups, haptophytes and cryptomonads (Okamoto et al. 2009).
References
Burki, F., Y. Inagaki, J. M. Archibald, P. J. Keeling, T. Cavalier-Smith, A. Horak, M. Sakaguchi, T. Hashimoto, D. Klaveness, K. S. Jakobsen, J. Pawlowski, and K. Shalchian-Tabrizi. 2009. Early evolution of eukaryotes: two enigmatic heterotrophic groups are related to photosynthetic chromalveolates. Genom. Biol. Evol. 1:231-238.
Cavalier-Smith, T. 1999. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J. Eukaryot. Microbiol. 46:347-366.
Cavalier-Smith, T., and S. von der Heyden. 2007. Molecular phylogeny, scale evolution and taxonomy of centrohelid heliozoa. Mol Phylogenet Evol 44:1186-1203.
Cuvelier, M. L., A. Ortiz, E. Kim, H. Moehlig, D. E. Richardson, J. F. Heidelberg, J. M. Archibald, and A. Z. Worden. 2008. Widespread distribution of a unique marine protistan lineage. Environ Microbiol 10:1621-1634.
Hackett, J. D., H. S. Yoon, S. Li, A. Reyes-Prieto, S. E. Rummele, and D. Bhattacharya. 2007. Phylogenomic analysis supports the monophyly of cryptophytes and haptophytes and the association of rhizaria with chromalveolates. Mol. Biol. Evol. 24:1702-1713.
Hampl, V., L. Hug, J. W. Leigh, J. B. Dacks, B. F. Lang, A. G. Simpson, and A. J. Roger. 2009. Phylogenomic analyses support the monophyly of Excavata and resolve relationships among eukaryotic "supergroups". Proc Natl Acad Sci U S A 106:3859-3864.
Keeling, P. J. 2009. Chromalveolates and the evolution of plastids by secondary endosymbiosis. J Eukaryot Microbiol 56:1-8.
Kim, E., A. G. Simpson, and L. E. Graham. 2006. Evolutionary relationships of apusomonads inferred from taxon-rich analyses of 6 nuclear encoded genes. Mol Biol Evol 23:2455-2466.
Klaveness, D., K. Shalchian-Tabrizi, H. A. Thomsen, W. Eikrem, and K. S. Jakobsen. 2005. Telonema antarcticum sp. nov., a common marine phagotrophic flagellate. Int J Syst Evol Microbiol 55:2595-2604.
Nikolaev, S. I., C. Berney, J. F. Fahrni, I. Bolivar, S. Polet, A. P. Mylnikov, V. V. Aleshin, N. B. Petrov, and J. Pawlowski. 2004. The twilight of Heliozoa and rise of Rhizaria, an emerging supergroup of amoeboid eukaryotes. Proc Natl Acad Sci U S A 101:8066-8071.
Not, F., J. del Campo, V. Balague, C. de Vargas, and R. Massana. 2009. New insights into the diversity of marine picoeukaryotes. PLoS One 4:e7143.
Not, F., K. Valentin, K. Romari, C. Lovejoy, R. Massana, K. Tobe, D. Vaulot, and L. K. Medlin. 2007. Picobiliphytes: a marine picoplanktonic algal group with unknown affinities to other eukaryotes. Science 315:253-255.
Okamoto, N., C. Chantangsi, A. Horak, B. S. Leander, and P. J. Keeling. 2009. Molecular phylogeny and description of the novel katablepharid Roombia truncata gen. et sp. nov., and establishment of the Hacrobia taxon nov. PLoS One 4:e7080.
Okamoto, N., and I. Inouye. 2006. Hatena arenicola gen. et sp. nov., a katablepharid undergoing probable plastid acquisition. Protist 157:401-419.
Okamoto, N., and I. Inouye. 2005. A secondary symbiosis in progress? Science 310:287.
Patron, N. J., Y. Inagaki, and P. J. Keeling. 2007. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr. Biol. 17:887-891.
Rice, D. W., and J. D. Palmer. 2006. An exceptional horizontal gene transfer in plastids: gene replacement by a distant bacterial paralog and evidence that haptophyte and cryptophyte plastids are sisters. BMC Biol. 4:31.
Sakaguchi, M., T. Nakayama, T. Hashimoto, and I. Inouye. 2005. Phylogeny of the Centrohelida inferred from SSU rRNA, tubulins, and actin genes. J Mol Evol 61:765-775.
Shalchian-Tabrizi, K., W. Eikrem, D. Klaveness, D. Vaulot, M. A. Minge, F. Le Gall, K. Romari, J. Throndsen, A. Botnen, R. Massana, H. A. Thomsen, and K. S. Jakobsen. 2006. Telonemia, a new protist phylum with affinity to chromist lineages. Proc Biol Sci 273:1833-1842.
Shalchian-Tabrizi, K., H. Kauserud, R. Massana, D. Klaveness, and K. S. Jakobsen. 2007. Analysis of environmental 18S ribosomal RNA sequences reveals unknown diversity of the cosmopolitan phylum Telonemia. Protist 158:173-180.
Vaulot, D., W. Eikrem, M. Viprey, and H. Moreau. 2008. The diversity of small eukaryotic phytoplankton (< or =3 microm) in marine ecosystems. FEMS Microbiol Rev 32:795-820.
Vørs, N. 1992. Ultrastructure and autecology of the marine, heterotrophic flagellate Leucocryptos marina (Braarud) Butcher 1967 (Katablepharidaceae/Kathablepharidae), with a discussion of the genera Leucocryptos and Katablepharis/Kathablepharis. Eur. J. Protistol. 28:369–389.
Title Illustrations

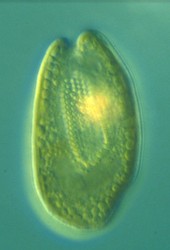
| Scientific Name | Cryptomonas sp. |
|---|---|
| Comments | from a freshwater sample. Cryptomonas lundii/borealis relationship. |
| Image Use |
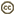 This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial License - Version 3.0.
|
| Copyright |
© 2008 Kerstin Hoef-Emden

|
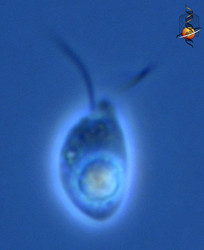
| Scientific Name | Telonema |
|---|---|
| Location | material from Limulus-ridden sediments at Plum Island, Massachusetts coast, USA |
| Comments | This cell has a food particle inside. |
| Source | Telonema |
| Source Collection | Micro*scope |
| Image Use |
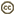 This media file is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike License - Version 3.0.
|
| Copyright |
© 2001 David J. Patterson

|
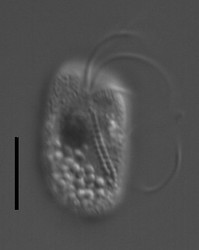
| Scientific Name | Katablepharis remigera |
|---|---|
| Location | Ucluelet, Vancouver Island |
| Comments | scale bar = 10 µm |
| Specimen Condition | Live Specimen |
| Identified By | Noriko Okamoto |
| Behavior | Swimming |
| Image Use |
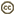 This media file is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike License - Version 3.0. This media file is licensed under the Creative Commons Attribution-NonCommercial-ShareAlike License - Version 3.0.
|
| Copyright |
© 2009

|
About This Page
This page is being developed as part of the Tree of Life Web Project Protist Diversity Workshop, co-sponsored by the Canadian Institute for Advanced Research (CIFAR) program in Integrated Microbial Biodiversity and the Tula Foundation.
Patrick Keeling

The University of British Columbia, Vancouver, British Columbia, Canada
Correspondence regarding this page should be directed to Patrick Keeling at
Page copyright © 2009 Patrick Keeling
 Page: Tree of Life
Hacrobia .
Authored by
Patrick Keeling.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
Page: Tree of Life
Hacrobia .
Authored by
Patrick Keeling.
The TEXT of this page is licensed under the
Creative Commons Attribution-NonCommercial License - Version 3.0. Note that images and other media
featured on this page are each governed by their own license, and they may or may not be available
for reuse. Click on an image or a media link to access the media data window, which provides the
relevant licensing information. For the general terms and conditions of ToL material reuse and
redistribution, please see the Tree of Life Copyright
Policies.
- First online 28 October 2009
- Content changed 28 October 2009
Citing this page:
Keeling, Patrick. 2009. Hacrobia . Version 28 October 2009 (under construction). http://tolweb.org/Hacrobia/124797/2009.10.28 in The Tree of Life Web Project, http://tolweb.org/






 Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site
Go to quick links
Go to quick search
Go to navigation for this section of the ToL site
Go to detailed links for the ToL site